This section describes how to perform a multi-dimensional reverse search [Fukuda2004, Waite2014, Waite2015] over N-best lists. Quoting from the abstract of [Waite2015], multi-dimensional MERT is
... a polynomial-time generalisation of line optimisation that computes the error surface over a plane embedded in parameter space. The description of this algorithm relies on convex geometry, which is the mathematics of polytopes and their faces.
Chapters 5 and 6 of [Waite2014] give a complete presentation of the underlying theory. This tutorial reviews the procedure used to carry out the two-dimensional MERT described in Section 4.2 of [Waite2015] and Section 6.4 of [Waite2014]. This second reference also explains some of the steps that are carried out within the procedure.
For this example we use the reverse search implementation of C. Weibel [Weibel2010]. Because his implementation has a GPL license it must be downloaded and compiled separately. The version used in the following example is 1.6.2f. The scripts used in the example assume that the shell variable $MINK_SUM points to the bin directory of this tool.
The following script will perform a mulit-dimensional MERT over 150 sentences. N.B. Before running reverse search it is necessary to generate the N-Best lists. To do this, follow Steps 1, 2, and 3 in the tutorial section MERT (Features Only) with M=150, and then run
> scripts/reverse_search.sh
Let us examine the key steps in this script.
Step 1. Feature Projection
Multi-dimensional MERT casts the line optimisation procedure as the affine projection of a linear space to a lower dimensional affine subspace. Within this lower dimensional affine subspace it is possible to use the reverse search algorithm to enumerate each set of hypotheses that can generated from the N-best lists. For this example a 3-dimensional affine subspace is constructed where the first feature dimension is the affine component, and the other two features map to the second f_1(e) and fourth f_3(e) features in the feature vectors F(e) contained in the N-best lists. The projection is performed as described in [Waite2014, Section 6.4]. The initial parameter value, 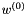 is the 12-dimensional parameter vector used to generate the N-best list (see
is the 12-dimensional parameter vector used to generate the N-best list (see configs/CF.mert.hyps and Applying Weight Vectors in Translation).
A 3x12 dimensional matrix 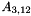 is formed from
is formed from 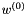 as described in [Waite2014, Section 6.4] so that any 12-dimensional feature vector
as described in [Waite2014, Section 6.4] so that any 12-dimensional feature vector 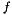 can be projected into the subspace as
can be projected into the subspace as 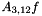 . This is done using the following script:
. This is done using the following script:
> zcat output/exp.mert/nbest/VECFEA/1.vecfea.gz | ./scripts/affine_project.py $FW [ [[-48.2233,-10.8672,16.0000],[-47.3144,-12.8613,17.0000],[-50.7771,-9.6670,16.0000], ...
The output format is required by the reverse search implementation and consists of 3-dimensional row vectors. In this example there are 100 vectors, one for each entry in the 100-best list. The reverse search algorithm requires a well known starting parameter to begin its search, and the implementation uses the parameter ![$[1,1,1]$](form_33.png) as its starting point. However we wish the search procedure to start from the projection of
as its starting point. However we wish the search procedure to start from the projection of 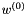 . To enforce this, the affine projection script subtracts 1 from the second and fourth parameter to force the reverse search implementation to use the starting parameter
. To enforce this, the affine projection script subtracts 1 from the second and fourth parameter to force the reverse search implementation to use the starting parameter [1,0,0]. The affine projection script also negates the feature vectors because the convention of reverse search is to maximise scores, as opposed to the cost minimization convention of HiFST.
Step 2. Convex Hulls of Projected Features
The following line computes the convex hull from the set of projected feature vectors. This is performed separately for each N-Best list:
> SETSIZE=150 > for i in `seq 1 $SETSIZE`; do cat output/exp.mert/polytope/$i.txt | $MINK_SUM/convexHull -d > output/exp.mert/hull/$i.txt;done
The result is a filtered set of feature vectors that form vertices of the convex hull for each sentence. Since any input feature vector that is an interior point is discarded, these files are shorter than the original N-best lists. These vertices are then compiled into an single file for input into the reverse search tool.
Step 3. Reverse Search
The reverse search is performed as follows:
> mkdir -p output/exp.mert/reverse_search > cd output/exp.mert/reverse_search > $MINK_SUM/minkSumForkGrid -c -n 12 < ../minksumin.txt
The -c option instructs the reverse search tool to compute the full normal cone associated with each result. This option can be omitted for faster computation.
The -n 12 option is the number of child processes to be used for parallel computation. In this case 12 are spawned.
The output of the tool is contained in the directory output/exp.mert/reverse_search. In this directory is an output file for each child process. Each line is these output files corresponds to a normal cone. Let us examine one line from an output file
[13,23,19,16,6,19,14,16,9,22,15,16,10,16,14,4,11,10,16,9,1,11,12,12,16,15,8,1,6,12,0,20,13,7,5,8,3,26,11,17,9,9,7,19,15,7,6,10,19,2,17,8,7,8,10,14,6,9,18,11,6,9,12,5,10,22,21,17,4,1,13,7,21,13,17,7,11,6,11,12,15,15,19,14,15,30,20,9,9,5,15,14,10,16,5,21,12,8,4,6,18,16,8,9,10,15,10,8,8,12,2,13,20,3,9,23,12,17,8,10,12,17,19,9,20,7,16,12,10,13,9,9,11,18,23,21,18,14,12,12,15,11,19,22,15,20,11,13,1,2] : [-12442.6,-1992.01,3084] : [38.187,303.022,-2189.79] : [[-0,-14.3453,10.706,1],[-0,8.69,-8.3183,-1],[-0,1.568,-0.1943,-0],[-0,-1.2371,0.1592,-0]]
This line is divided into four fields separated by the colon character :
- The first field represents the set of hypotheses maximised by the parameters in this cone. It is a vector of 150 elements, with each element corresponding to an index to a vertex in the convex hull associated with each input sentence
- The second field is the sum of all the vertices
- The third field is a parameter interior to the normal cone which maximises all the vertices in the first field
- The fourth field is an optional field associated with the
-coption. It is the set of rays that define the cone
The parameter vector (the third field) can be used to rescore the N-Best lists by their projected feature vectors: each parameter vector will return the hypotheses identified by the first field (barring numerical errors).
Due to the geometry of a normal cone, the parameter vectors can be mapped from 3 to 12 dimensions, as follows:
- Parameter vectors for which the first element of the parameter vector is negative are rejected
- The parameter vector is scaled by the first element (
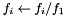 )
) - A new 12-dimensional parameter vector is created by adding the second and third element of the scaled projected feature vector to the second and fourth elements of the initial parameter
Step 4. Reranking
Once the reverse search is complete, we should complete a sanity test to ensure that the parameters found by the reverse search correctly rerank the N-best lists.
The first step is to map the vertices in the convex hull to the feature vectors in the original N-best list.
> scripts/map_vertices.py output/exp.mert/polytope output/exp.mert/hull > output/exp.mert/mapping.txt
The result of this script is a pickled python map object that provides the mapping between index of a vertex in a convex hull to the index of the corresponding feature vector in the N-best list. Using this mapping between convex hull and the indices of the N-best list, the sanity test is executed by the following script:
> cat output/exp.mert/reverse_search/result.* |\ scripts/reverse_search_test.py output/exp.mert/mapping.txt output/exp.mert/nbest/VECFEA $GW > log/log.reverse.search.sanity
The 3-dimensional parameter vectors found by the reverse search are mapped up to 12 dimensions (as described above) and used to re-rank the 150 N-best lists. The index of the top scoring hypotheses of the reranked lists should match the mapped index from the first field of the reverse search output. If the indices do not match, the sanity test script prints a line of the form:
Discrepancy! For input sentence 31 featue vector 2 is the top scorer
Due to floating point errors there will be some N-best lists that are reranked incorrectly. For reference, using 150 input sentences with 100-best lists generated 346,526 parameters. Of these parameters 185,475 were accepted for testing (i.e. had 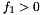 ), which means that 27,821,250 (=150 * 185,475) reranking tests were performed. Out of these tests 28,282 rerankings had the wrong top scoring hypothesis, which is approximately 1% of all the reranking tests performed.
), which means that 27,821,250 (=150 * 185,475) reranking tests were performed. Out of these tests 28,282 rerankings had the wrong top scoring hypothesis, which is approximately 1% of all the reranking tests performed.
